Decoding the molecular mechanisms controlling T cell development and activation.
T cells play a crucial role in our immune system, responding to threats by recognizing specific parts of pathogens. This recognition triggers a chain of signals inside the cell, leading to different immune responses.
Given the essential role of T cells, tremendous efforts have been made over the last decades to map and characterize the molecular mechanisms responsible for signal propagation inside T cells.
The intracellular signals controlling T cell activation are mediated by hundreds of molecular components that are highly regulated and may interact with each other. To tackle this complexity, it is important to develop integrated strategies to gain a comprehensive understanding of the molecular regulations that control cellular functions, and hence immune responses.
Romain Roncagalli and his team aim to understand the molecular mechanisms and cellular processes that determine the fate of T cells under various physiological conditions.
For years, the analysis of T cell signaling pathways has been limited by the experimental tools available, and often restricted to a limited number of proteins selected based on prior knowledge. Recent technological advances now allow to characterize protein networks and determine the abundance and post-translational regulation of almost each protein expressed in a cell. Our team develops original Mass Spectrometry (MS)-based approaches to generate comprehensive and dynamic models of the molecular processes that are associated with T-cell’s states.
Using genetically modified mouse models, we look at how proteins interact and change when T cells encounter different signals. In particular, we seek to characterize the stoichiometry and dynamics of protein-protein interactions (PPIs) encoding the engagement of the TCR and others receptors expressed on the cell surface of lymphocytes.
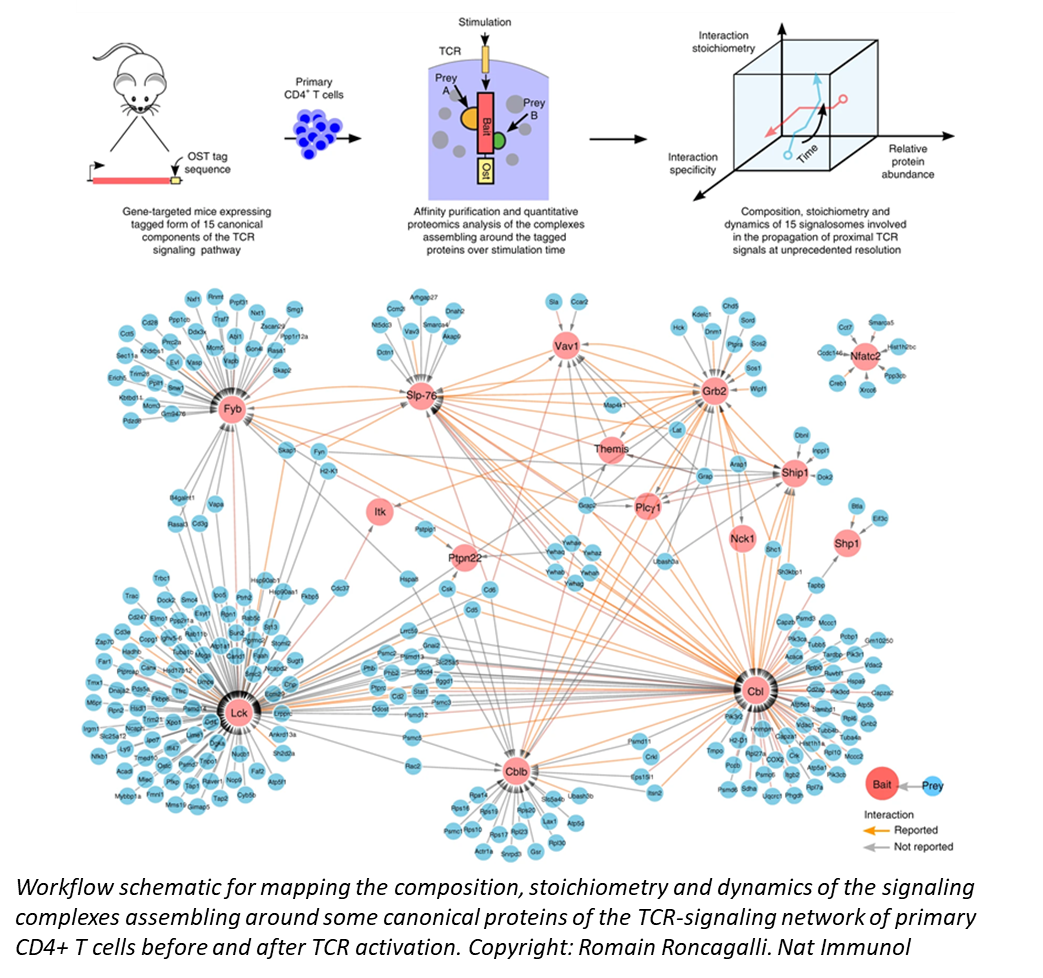
Using time-resolved high-resolution phosphoproteomics, we are also able to identify, quantify, and characterize the phosphorylation dynamics of thousands of phosphorylation sites in primary T cells after TCR stimulation. Our work reveals a coherent orchestration of biological processes and functional modules associated with cytoskeletal remodeling, transcription, translation, and metabolic processes in the early stages of TCR engagement. Using similar approaches, we also explore the molecular mechanisms that enables T cells to discriminate antigens of varying affinities.

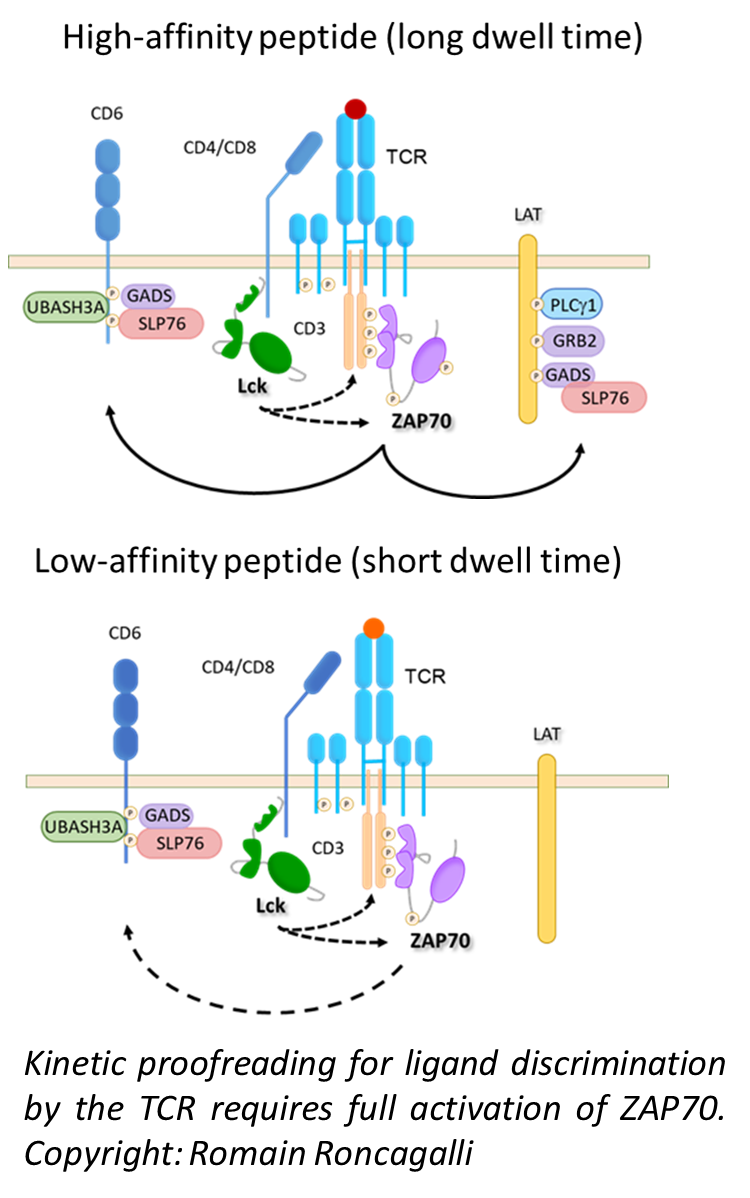
In the course of our research, we have come to realize that the mechanisms regulating T cell signaling, and therefore their fate, are far more complex than originally thought, and that many molecular components essential to T cell regulation have not yet been characterized. Our aim is to assess this complexity at multiple scales, from the molecule, through the cell, to the living organism. Indeed, our research strategy consists in going back and forth between high-throughput methods and targeted molecular analysis to associate molecular regulations with cellular phenotypes that emerge under different physiological conditions.