|
|
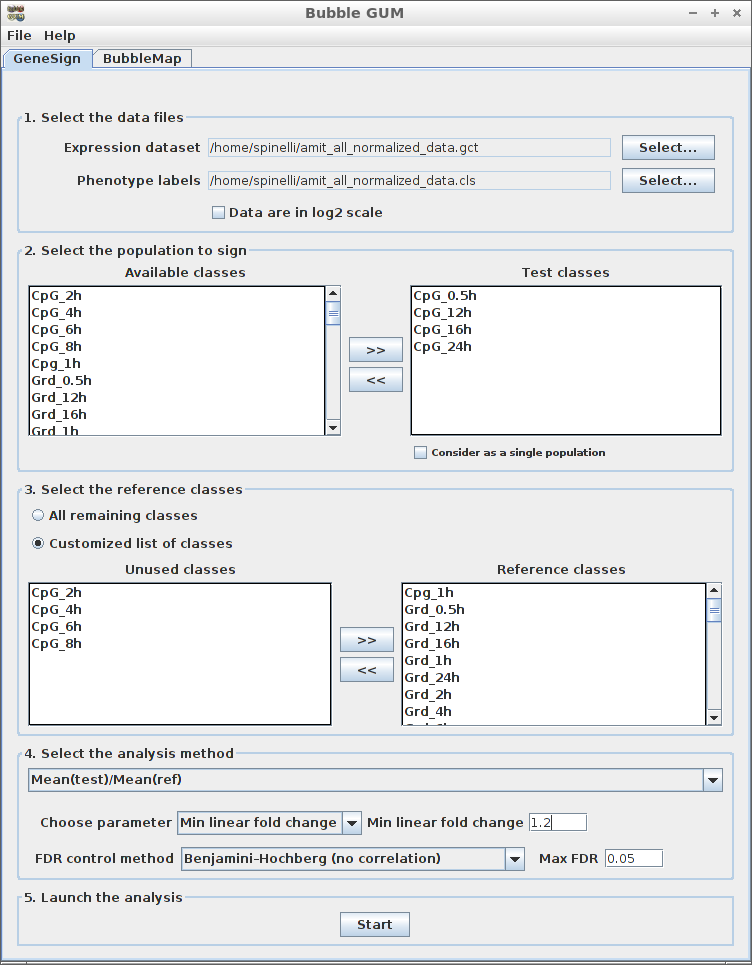
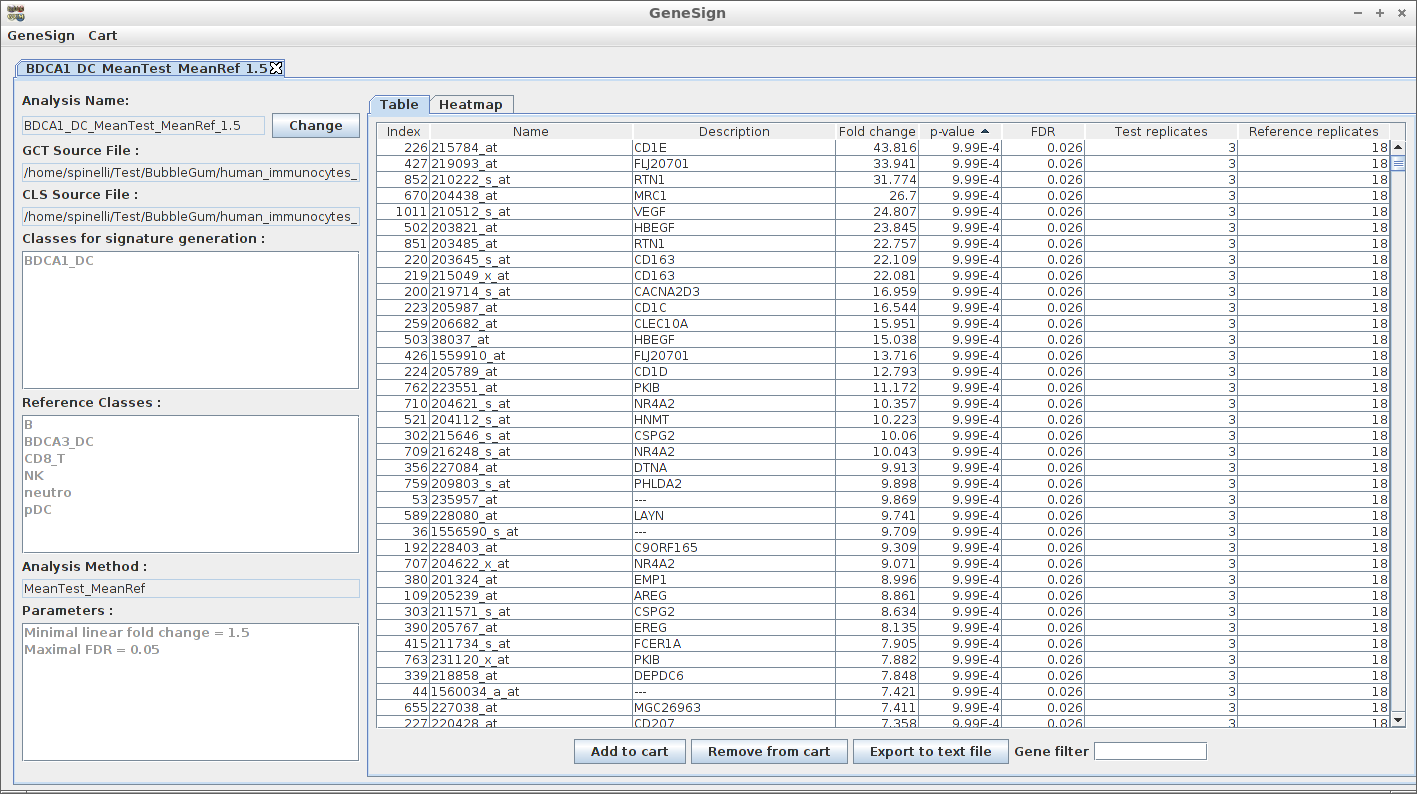
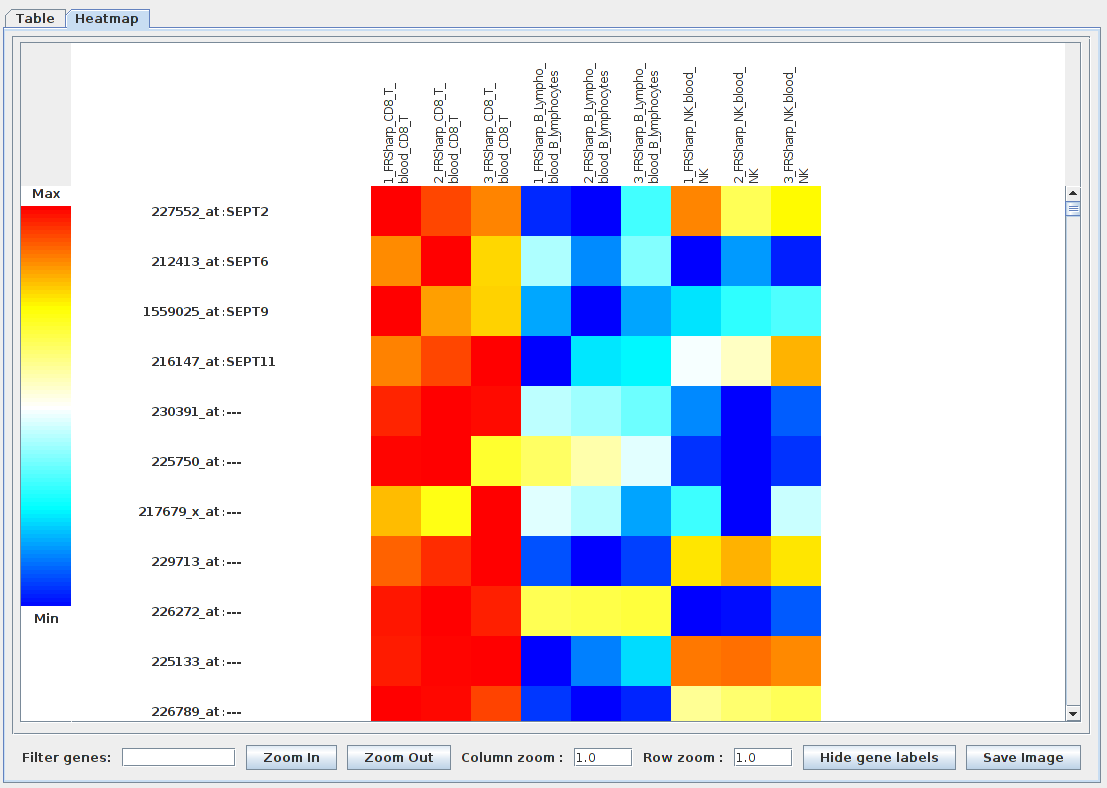
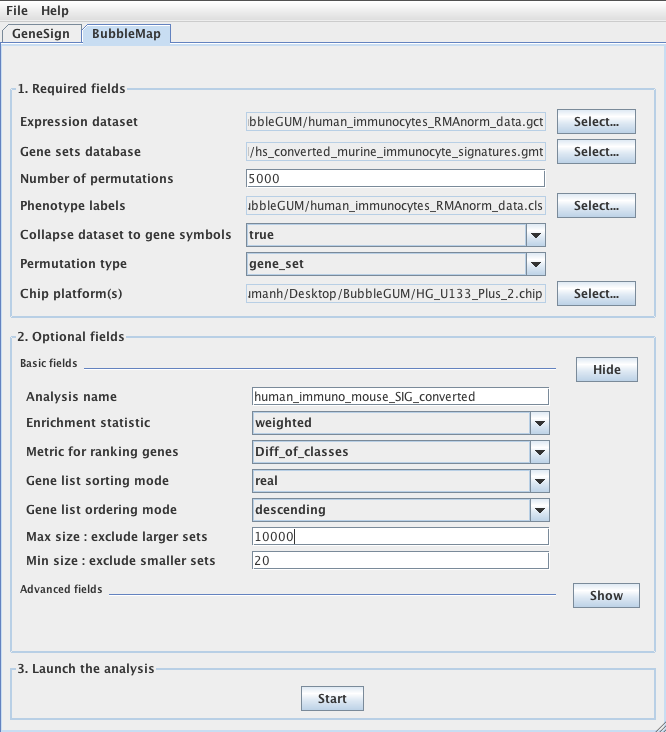
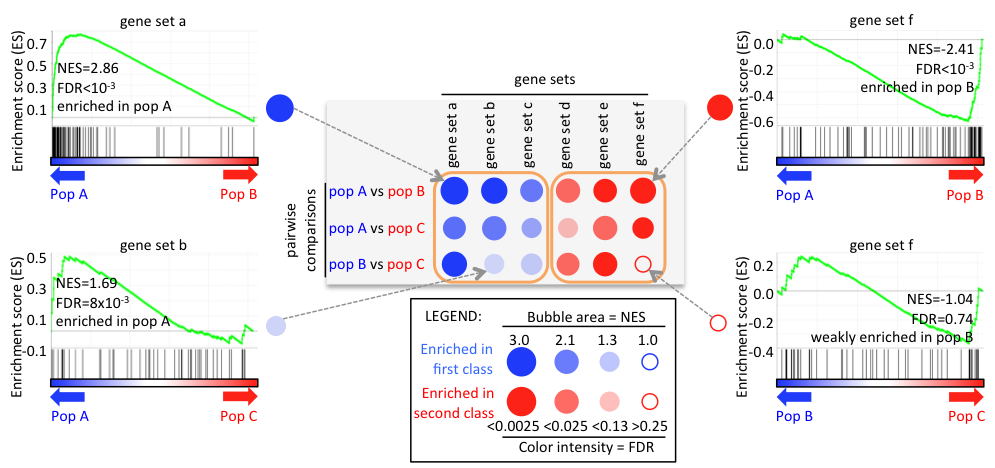
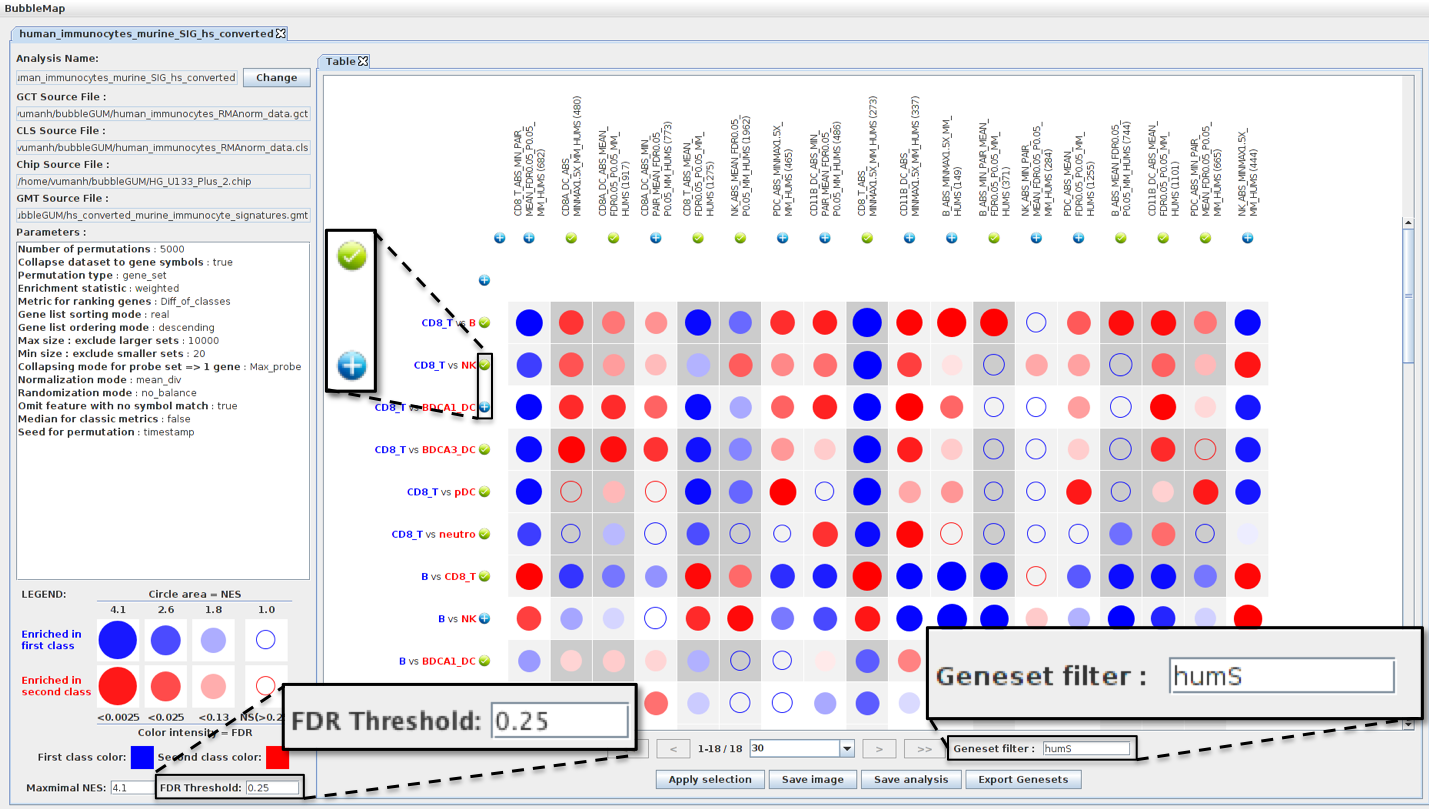
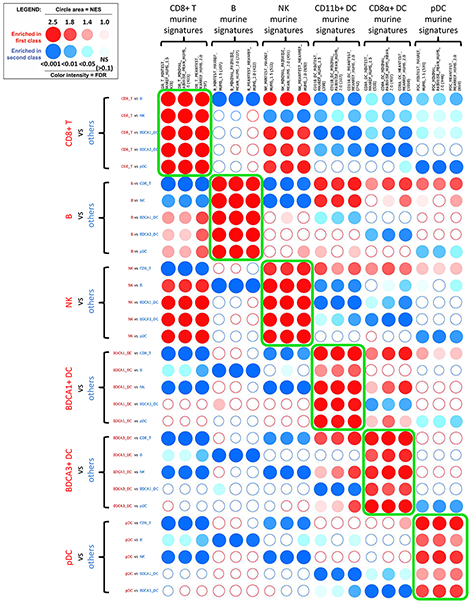
Recent advances in the analysis of high-throughput expression data have led to the development of tools that scaled-up their focus from the single-gene level to the gene set level, as for example the popular Gene Set Enrichment Analysis (GSEA) algorithm. GSEA detects modest but coordinate regulations of groups of presumably related genes between two sets of microarrays, in a pairwise comparison manner. Integrating multiple GSEA results is a crucial next step in an era where it is very frequent to cumulate expression data for multiple conditions in one study. Moreover, the power of gene set based analyses relies on the relevance of the gene sets. The ability to generate optimal homemade gene sets is therefore valuable. We present Bubble GUM (GSEA Unlimited Map), a computational tool that allows to automatically extract phenotype signatures based on transcriptomic data and to perform easily multiple GSEA runs in a row. The result is displayed at the global scale, as enrichment patterns or bubble maps, in order to help the interpretation of multiple interconnected enrichment plots. Bubble GUM aims to be used by both bioinformaticians and biologists, through an intuitive easy-to-use interface.
REFERENCE:
Spinelli L., Carpentier S., Montanana F., Dalod M., Vu
Manh TP.
CONTACT:
spinelli@ciml.univ-mrs.fr
vumanh@ciml.univ-mrs.fr
REGISTER:
Since BubbleGUM uses GSEA developped by the Broad Institute of MIT and Harvard, don't forget to register to MSigDB before you start enjoying Bubble GUM software:
http://www.broadinstitute.org/gsea/login.jsp
Download BubbleGUM
February 19th, 2016 : New version 1.3.19 - See release notes
|
|
|
For Linux, Mac and Windows versions, download the zip file and uncompress it in some location. Then double-click on the
file corresponding to the level of memory you want to allocate : Normal (run_4GB) or High (run_8GB).
Note: You may need to activate the "execute" property on the files in order to run them.
Documentation and sources
|
|
Demo files
|
|
|
|
|
|
|
|
|
|
|
|
|
|