Dendritic cells (DCs) form a family of cells endowed with the unique capacity of stimulating naive T cells and orchestrating primary immune responses. Our team is specialized in the cell biology of this important cell family with both innate and adaptive immune functions.
We are therefore expert at isolating or differentiating in vitro different lineages of both mouse and human DCs. Our research focus on deciphering the whole of biochemical changes occurring during cellular activation by microbes or stress, and how misregulation of these molecular pathways contribute to chronic inflammation, autoimmunity or cancer. Our work provides a novel framework to develop therapeutic strategies to modulate the immune response during disease or vaccination.
Overview
Our aim is to elucidate the molecular mechanisms by which DCs and other cells control antigen processing and cytokine production, particularly during microbe detection or upon stress exposure. In the recent years, We have been focusing on mRNA translation as well as protein synthesis quality control, and their mutual relationship with the Integrated Stress Response (ISR), Autophagy and MHC restricted antigen presentation. We are also characterizing specific molecules that regulates membrane traffic and controls conventional DC and type-I interferon producing plasmacytoïd DC function.
The laboratory is specialized in interdisciplinary approaches using cell biology and immunology techniques together with advanced cell imaging, multiscale omics and mathematical modelling to try to meet the challenge of understanding the massive molecular changes occurring upon microbe detection by mammalian cells and whole mice.
Our laboratory has accumulated an extensive scientific know-how in the study of DCs, and repetitively proven that it is able to transform innovative concepts in coherent projects that are all integrated in one global experimental strategy (Claudio et al., 2013, Arguello et al., 2015, 2016; Dalet et al., 2015, Reverendo et al., 2018, Valečka et al., 2018).
Our ambition is to explore uncharted research areas, bringing a broader understanding of the essential biochemical functions required for the acquisition by DCs of their unique immune-regulatory functions and propose new therapeutic alternatives for the control of immuno-dependent diseases.
Our team is co-managed by Philippe Pierre (DR1,CNRS) and Evelina Gatti (DR2, CNRS), who is responsible of the membrane traffic projects performed in the laboratory and coordinates the CNRS Laboratoire International Associé (LIA) “MISTRA” with the Aveiro University in Portugal (http://www.ua.pt/ibimed/page/21080).
eEF2AK, stress and microbial detection
Translation initiation factor 2-alpha (eIF2) phosphorylation strongly attenuates mRNA translation initiation and activates several gene expression programs collectively known as the Integrated Stress Response (ISR). To date, four eIF2 kinases (eEF2AK) are known to mediate eIF2 phosphorylation: PKR, PERK, GCN2 and HRI. We have demonstrated that DCs, at steady state both in vitro and in vivo, display unusually high levels of phosphorylated-eIF2 characteristics of “stressed” cells.
DC activation by microbe-associated molecular patterns (MAMPs) induces eIF2 de-phosphorylation through the expression of the phosphatase-1 co-factor, GADD34, also known, as PPP1R14A. Interestingly, the rapid translational arrest, normally mediated through eIF2 phosphorylation in response to stress (including dsRNA recognition by PKR and ER stress by PERK) does not occur in activated DCs (Clavarino et al., 2012a). GADD34 expression in DCs and in mouse embryonic fibroblasts (MEFs) is however absolutely required for the normal production of IFN-ß in response to dsRNA (e.g. poly I:C) and is thus an integral part of the anti-viral response (Clavarino et al., 2012b). In MEFs, GADD34 expression is completely dependent on transcription factor IRF3 and is co-regulated with IFN-ß transcription. IRF3-dependent transcription explains therefore the critical requirement for GADD34 in protecting mouse neonates from Chikungunya virus infection (Clavarino et al., 2012b). GADD34 and associated signaling cascades were shown, to be important for cytokine production in DC and other immune cells exposed to MAMPs (Clavarino et al., 2012b). Based on these results, we have developed several mathematical models that demonstrate the oscillating nature of protein synthesis and the role of the GADD34 specific translation regulation in the stochastic appearance of type-I IFN production observed upon nucleic acid detection in different cell types.
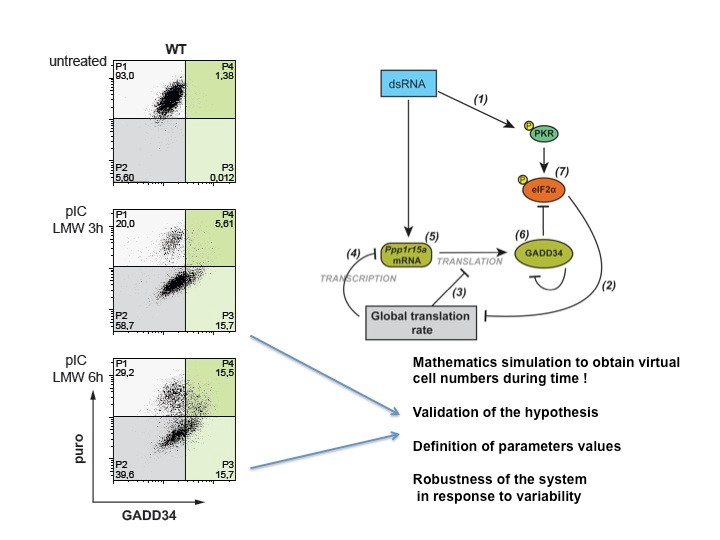
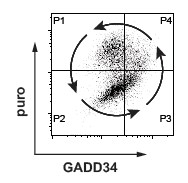
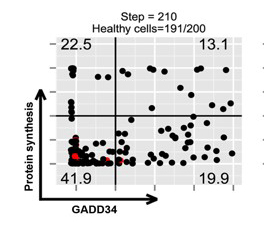
Intracellular FACS measurement of translation using SUnSET after dsRNA stimulation of MEFs, demonstrate that cells undergo translation arrest (P2) prior exiting this inhibition through GADD34 expression (P3, P4) and produce type-I IFN following a specific cycle (P4). This situation was mathematically modeled during virtual time and shown to be partly responsible for stochastic IFN production (Dalet el al. 2017).
GADD34 represents therefore an attractive target for limiting cytokines release in interferonopathies like systemic lupus erythematosus (SLE). Pharmacological inhibition in vitro and in relevant mouse SLE models has turned to be beneficial to limit auto-immunity and cytokine shock underlining the anti-inflammatory potential of inhibiting GADD34/PP1A (Perego et al. 2017 and 2018).
Endocytic molecules trafficking and DC function
MAMPs detection impacts membrane organelles organization, and more particularly endocytosis to allow the efficient acquisition by DC of their immuno-stimulatory functions. Endosomes dynamic is also particularly important to regulate the capacity of nucleic acids-activated TLRs such as TLR3 or TLR9 to trigger different signaling pathways leading either to type-I IFN production or pro-inflammatory cytokines secretion. We were fist to demonstrate that MARCH I, a membrane associated RING-CH ubiquitin E3 ligase, promotes in DC, the ubiquitination of peptide-loaded MHC II, inducing their internalization from the cell surface, and thus impairing antigen presentation in the absence of an appropriate stimuli (De Gassart et al., 2008). We have recently focused on MARCH9, the only known MARCH ligase family member up-regulated by MAMPs. We found that MARCH9 is partly responsible for MHC I and CD1a ubiquitination and that in the absence of MARCH9, MHC I molecules are retained in the Trans Golgi Network (TGN), instead of accumulating in a syntaxin 6-positive specialized early endosomes (Rigotti et al., 2017). Collectively our results suggest that MARCH9-mediated ubiquitination in the TGN, targets a fraction of MHC I to specific endosomes involved with antigen cross-presentation (Rigotti et al., 2017).
We have identified a new member of the lysosome-associated membrane protein (LAMP) protein family, the brain and DC associated LAMP-like molecule (BADLAMP/LAMP5), as a specific marker of human plasmacytoïd DCs (pDCs) and blastic pDC neoplasms (BPDCN) (Defays et al. 2011). Further characterization indicated that in most species, BAD-LAMP expression is confined to neurons in the forebrain and olfactory bulb, where it fine tunes the dynamics of evoked GABAergic transmission (collaboration with H. Cremer laboratory at IBDM, Marseille, Tiveron et al., 2016). Our recent work on BAD-LAMP function in human pDC, indicates that it is a specific chaperone/regulator of Human TLR9 traffic and signaling in endosomes, setting the threshold of pDC activation by nucleic acids during cancer and infection (Combes et al., 2017). We have also observed rapid and transient reorganization of the endosomal network of pDCs upon nucleic acid detection.
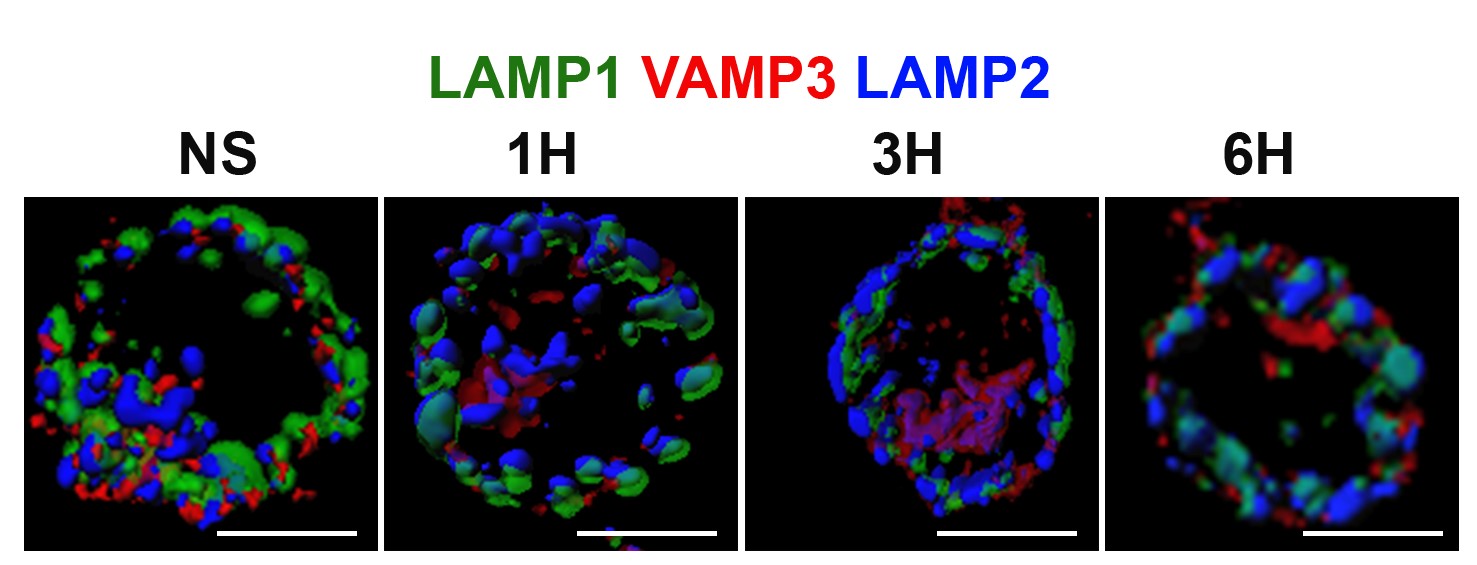
Different endosomal markers are re-organized upon pDC activation. IMARIS 3D reconstruction of confocal microscopy images (Combes et al. 2017).
Role of autophagy in antigen presentation and protein aggregation
During our previous studies on dendritic cell aggresome-like induced structures (DALIS) and translation regulation (Lelouard et al., 2002; 2004; 2007), we demonstrated that defective proteins (DRiPs) aggregation impacts MHC I restricted antigen presentation by controlling the flux of antigenic peptides in maturing DCs. We have further shown that DC activation by TLR ligands activates the mTOR pathway, which leads to an augmentation in bulk protein synthesis and decreases profoundly basal autophagic flux, resulting in the accumulation of poly-ubiquitinated antigens in DALIS (Terawaki et al., 2015). Autophagy describes different eukaryotic mechanisms of protein degradation, which result in the transfer to and the degradation of organelles or cytosolic material in the lysosomes. Given that autophagy also contributes to MHC II-restricted presentation of cytosolic antigens, TLR-mediated inhibition of autophagy incapacitates DCs to present endogenous and cytosolic antigens onto MHC II, with probable consequences on peripheral immune tolerance establishment. The expertise of our laboratory both in protein synthesis regulation and membrane traffic dynamic was absolutely key to carry on with these studies requiring many different technical skills including advanced light microscopy. DALIS, contain the p62 and NBR1 autophagy receptors and are only detected in activated DCs in vivo. DALIS are therefore markers of reduced autophagy and decreased endogenous MHC II restricted antigens presentation capacity in activated DCs (Wenger et al., 2012, Terawaki et al. 2015). Upon exploration of the consequences of different pathological immune conditions like asthma, we have demonstrated that long-term exposure to the cytokine IL-4, positively impacts DCs capacity to perform autophagy and increases considerably cytosolic antigens presentation on MHC II. At the molecular level, IL-4 exposure changes the status of the mTORC1 and GCN2 (EIF2AK4) pathway, rendering cells less sensitive to TLR-dependent autophagy inhibition, while inducing the expression of the RUN and FYVE domain-containing protein 4 (RUFY4) (Terawaki et al., 2015). RUFY4 was shown to interact with Rab7 and PLEKHM1 (McEwan et al., 2015), thus positively regulating autophagy and promoting lysosomes tethering. RUFY4 restricted expression to lung macrophages, neutrophils and CD11b+ DCs from house dust mite allergic mice suggests a particularly central role of this molecule for immune regulation and potentially the development of TH2 pathologies (Terawaki et al., 2015).
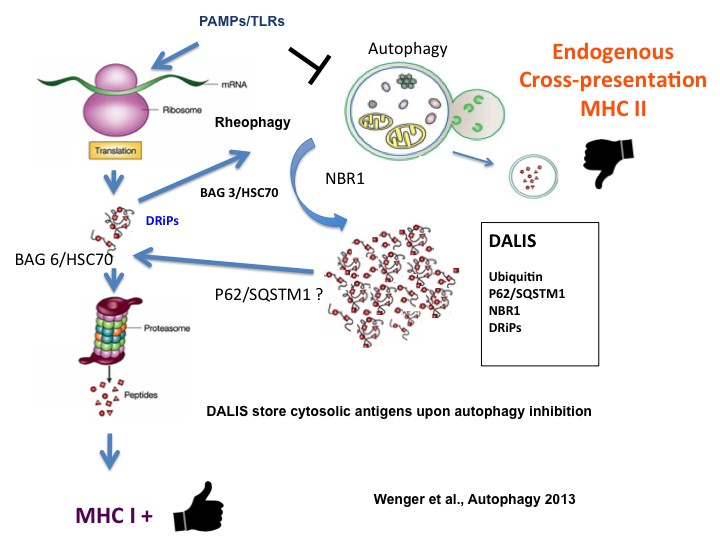
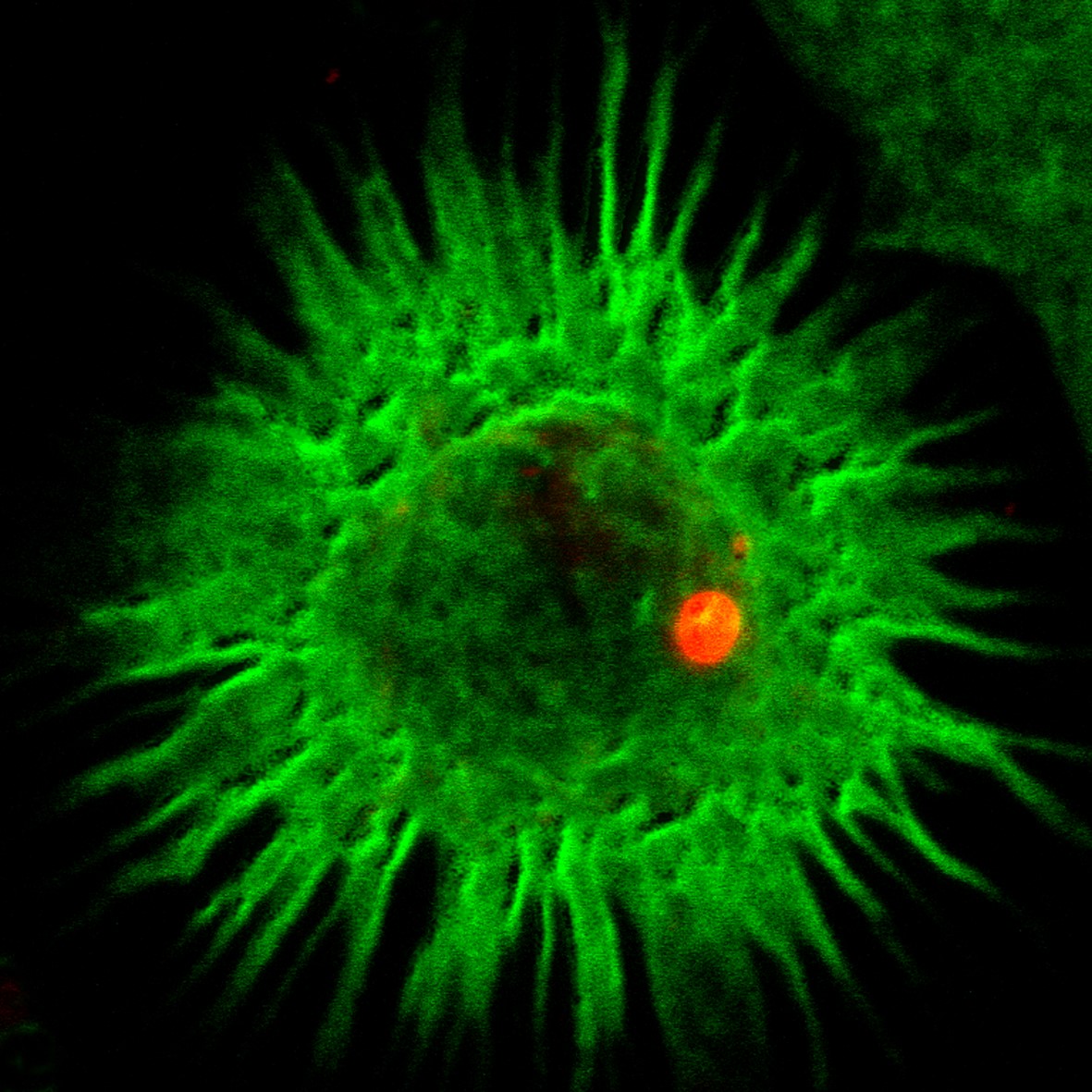
Left: Representation of DALIS formation in DCs and Drips re-targeting to proteasome during autophagy inhibition after PAMPs detection. Right: DALIS (ubiquitin in red) in maturing DCs (MHC II in green) (Terawaki et al., 2015). Copyright P Pierre, CIML.
System biology approaches and novel methodologies to monitor protein synthesis
Early during our work, We have been confronted to the necessity of performing gene expression and biochemical pathway analysis using structured ontologies. During the generation of DC-ATLAS a virtual tool to map DC signaling pathways (Cavalieri et al. 2011), we realized that it was particularly hard to get a picture of gene expression data in the context of the whole ontology at hand, and to easily interrogate it. We have developed Voronto (In coll. with R. Santamaria, U. Salamanca), a tool that integrates expression data and biological ontologies. Voronto allows the analyst to explore the whole ontology and detect changes on expression patterns using Voronoi tessellation (Santamaria and Pierre, 2012 ; Terawaki et al, 2015). Moreover, we extended our collection of molecular data on DC activation by performing organelle proteomics (In coll. with M. Desjardins, U. Montreal, Canada) and establishing late endosomes and lysosomes protein compositions from mouse bone-marrow derived DCs and macrophages (Duclos/Clavarino et al. 2012).
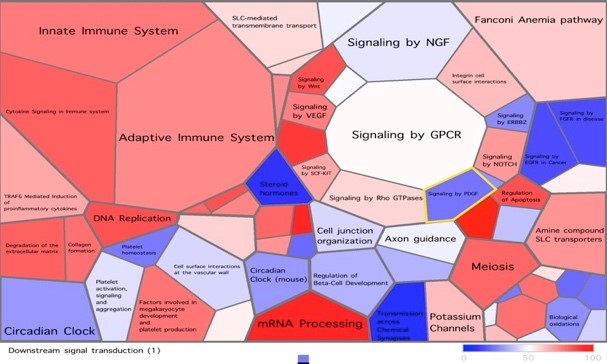
GEO visualization using tessellation of a gene expression array using Voronto. http://vis.usal.es/voronto/Intro.html
Our strong interest in protein synthesis regulation and misfolded proteins has also led to the invention of the “SunSET” and “SunRiSE” methods (Schmidt et al., 2009 and Argüello, et al. 2018), which are based on the incorporation of puromycin and its detection by cognate monoclonal antibodies to monitor protein synthesis. These techniques have been diffused world-wide and have led to more than 400 publications across many different biological research fields. This technology was necessary to propose the protein synthesis oscillation theory (Dalet et al., 2016) and demonstrate the existence of protein synthesis in the cell nucleus (David et al., 2013)
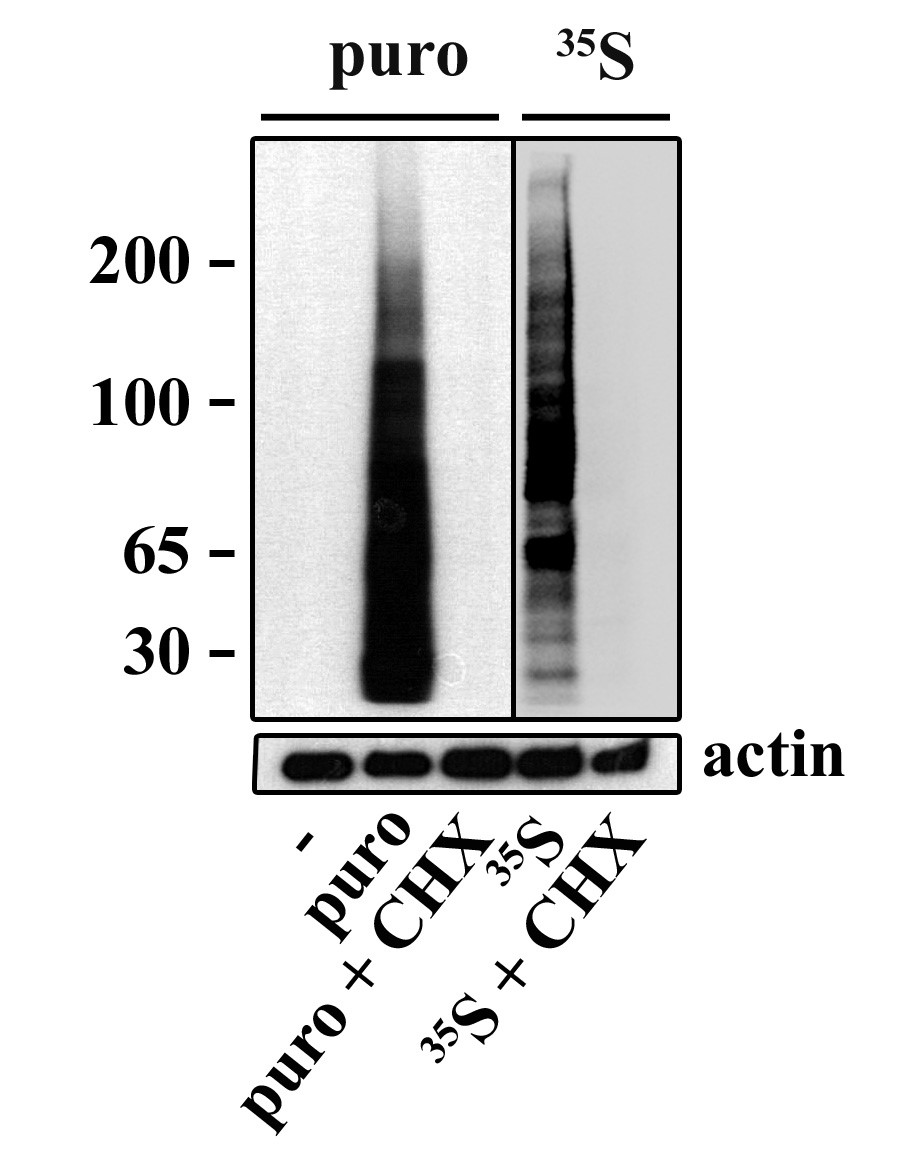
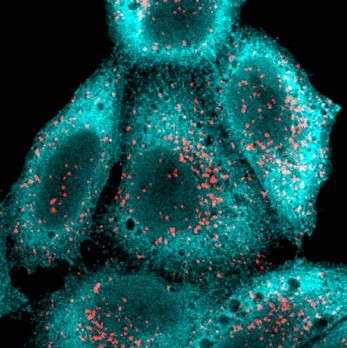
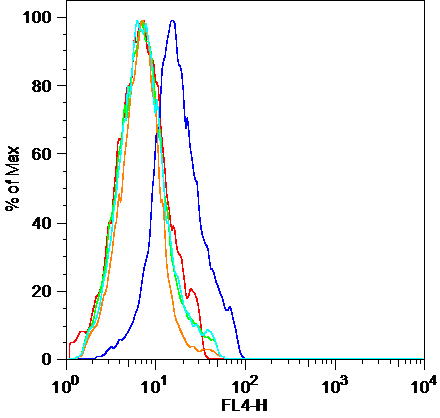
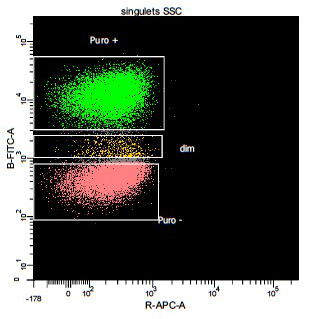
Puromycin incorporation can be used to monitor protein synthesis using mAb
Left) Detection and measurement of protein synthesis in NIH 3T3 cells using a puromycin pulse, compared to S35-methionine labelling. Center L) confocal immunofluorescence of HeLa cells pulsed with with puromycin (cyan) and co-labelled with lysosomes (red), shows ER bound translation activity. Center R) FACS detection of surface puromycin exposure (SUnSET) in NIH3T3 cells, untreated (blue line, right) or upon translation inhibition with cycloheximide (red line). Right) MEFs exposed to dsRNA and puromycin labelled. Protein synthesis arrest in about 50% of the cell population were detected using intra-cellular permeabilization and SUnSET detection, active cells are in green and inhibited in red for better visualization. (Schmidt et al. 2009; Argüello et al. 2018).
CNRS-University of Aveiro (PT) International Associated Laboratory “MISTRA”
This project represents a multidisciplinary and integrated effort to answer the question: “Is translation regulation hard wired into environmental responses to help cells evolve and adapt to stressful conditions?” We have observed translation-based responses, operating through dynamically reprogrammed tRNA modifications and distinct codon usage patterns in response genes, are essential to surviving stress. tRNA modifications and codon usage patterns are also linked to mistranslation, with protein errors promoting immediate phenotypic diversity and, notably, also DNA mutations. We propose that mistranslation driven by tRNA modifications and codon usage variations are programmed responses to stress and can induce stress responses. We have also shown that ER-stress and autophagy inhibition are impacting immunocytes and create pro-inflammatory responses. We carry-out our research using as a model dendritic cell activation and are currently assembling experimental information on different tRNA pathways to answer the question about the relationship between tRNA homeostasy and Immunity. This open wall international associated laboratory coordinated by Evelina Gatti is sponsored by CNRS, the Institute for Research in Biomedicine (iBiMED) of the University of Aveiro (https://www.ua.pt/ibimed/) and the Ilidio Pinho Foundation.