We investigate the molecular mechanisms involved in the control of differentiation events during lymphocyte development, including antigen receptor gene expression and recombination. One approach is to characterize cis-regulatory elements and bound nuclear or transcription factors at lymphoid gene specific loci and to infer their impact on chromatin structure using molecular biology approaches and, especially, high-throughput genomics (microarrays, ChIP-Chip, ChIP-seq, RNA-seq, MeDIP, etc…) and bio-informatics; as well as transgenic mouse and gene targeting (knockout/knockin) technologies.
In parallel, transgenic and mutant animals are being used to study precursor-product relationships along lymphoid cell developmental pathways, applying molecular and cellular biology approaches and mathematical simulation [in collaboration, notably, with the team ‘Non-Linear Dynamics’ lead by M. Pettini at the Center for Theoretical Physics (CPT), Luminy Campus].
We also analyze the effect of epigenetic events associated with oncogenic conversion and the process of leukemogenesis; or associated with the induction of central tolerance in the thymus (M. Irla). Hense, using knockin techniques, genomics and proteomics, we intend to define novel factors and the regulatory networks that sustain lymphocyte onto/oncogeny and full maturation.
Overview
Ongoing research projects: 1-3 and 5 are supervised by P FERRIER ; 4 by M. IRLA See bibliography...
1- Regulation of V(D)J recombination
V(D)J recombination, the DNA rearrangement process that assembles antigen receptor genes, is required for lymphoid cell development.
We explore the molecular mechanisms involved in the regulation of T cell receptor (TCR) gene recombination during T cell differentiation, focusing on the TCRβ gene enhancer (Eβ) that commands TCRβ gene expression through the lifespan of T lymphocytes. We expect insight into enhancer specific function(s) in adaptive immunity and TCR gene allelic exclusion.
2- Epigenetic dynamics of T-cell development and oncogenesis
Histone post-translational modifications shape the chromatin landscape. How these epigenetic tags exactly combine in order to regulate transcriptional activity during cell developmental processes is an open question.
We use primary thymocytes to investigate gene expression and epigenetic tagging by active (H3K4me1/2/3, H3K36me3) vs. repressive (H3K9me2, H3K27me3) chromatin marks, prior to and after the TCR selection checkpoints during T-cell differentiation.
Chromatin plays an important role in oncogenesis. Genome-wide approaches now allow large-scale characterization of epigenetic landscapes also in cancer cells.
In collaboration with researchers and clinicians from several Cancer Research Institutes and Hospitals, we define epigenetic signatures of Acute Lymphoblastic and Myeloid Leukemias and Lymphomas, with potential implications toward disease-oriented diagnosis, prognosis and therapy.
3- Basis of gene transcription during T-cell differentiation
The Carboxy-terminal-Domain (CTD) of RNA Polymerase (Pol) II harbors repetitions of a conserved heptapeptide that can be phosphorylated on Ser5 (S5P) during transcriptional initiation; on Ser2 (S2P) during elongation; and on Ser7 [S7P; characterized recently]. Likely, S5P/2P/7P combinatorial features play important roles in transcriptional controls. Enhancers may also influence the rate of transcriptional initiation via the recruitment of General Transcription Factors (GTFs) and Pol II prior to their translocation to the promoter sites.
We perform genome-wide profiling of GTFs and Pol II phosphorylated isoforms as well as various epigenetic marks, in differentiating primary T-cells. Our ChIPseq data shed light on poised areas of Pol II and the function of CTD isoforms during gene transcription. Notably, we observed Pol II and GTFs binding to known and novel enhancers in some cases resulting in local transcription. Clustering of TBP and Pol II-S5P plus RNAseq analysis indicated that the initiating transcriptional machinery spreads over large inter- and intragenic areas. We thus defined new regulatory genomic elements, which we called TIPs (Transcriptional Initiation Platforms).
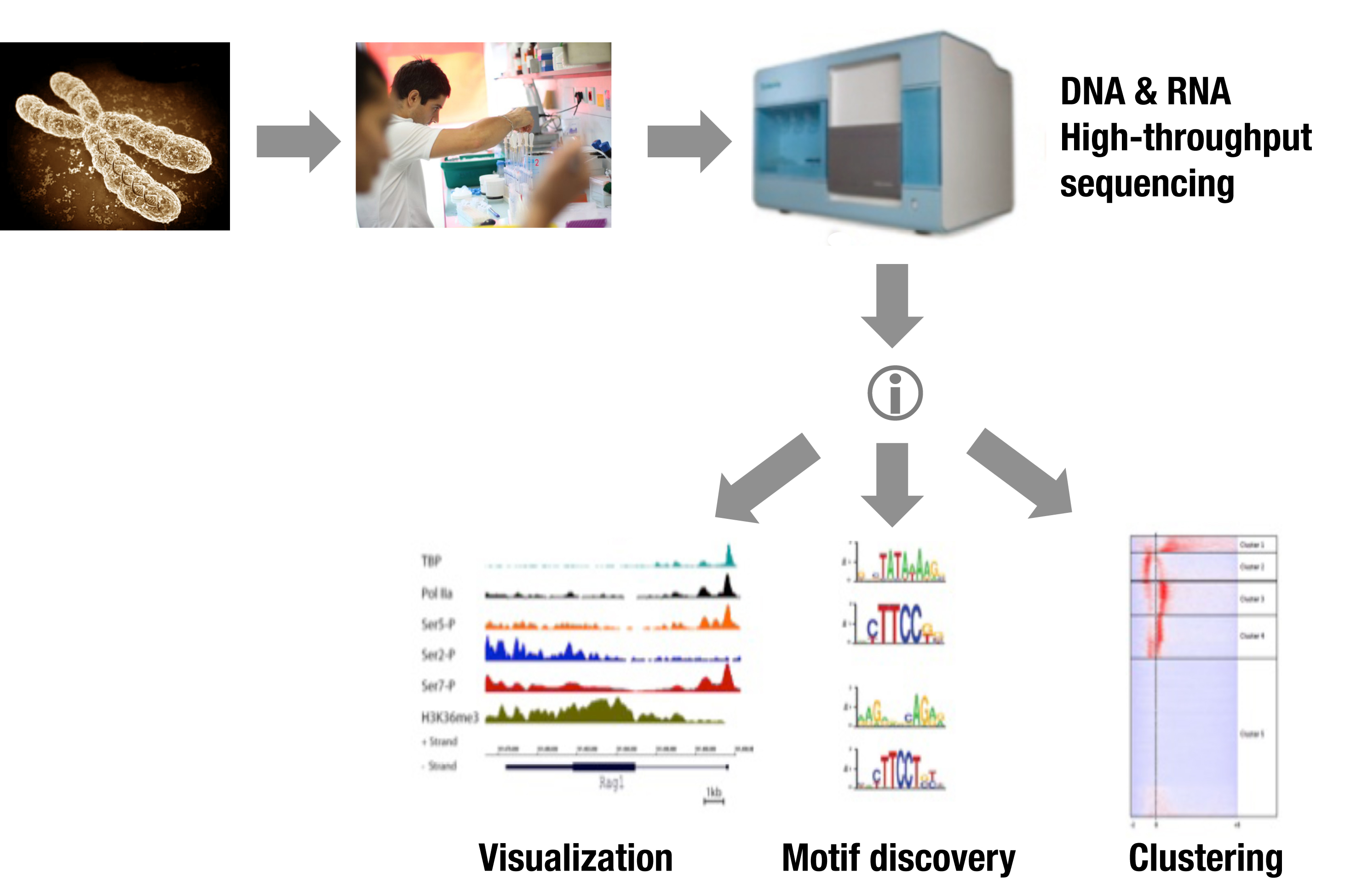 Epigenetics of Lymphoid Cell Development
Epigenetics of Lymphoid Cell Development
4- Mechanisms controlling the induction of central tolerance in the thymus
Healthy individuals mount immune responses directed against pathogens while avoiding autoimmune attacks on their own tissues. This ability to distinguish between self and non-self is referred to as immunological self-tolerance. The thymic medulla provides a specialized microenvironment dedicated for T-cell tolerance induction. Medullary thymic epithelial cells (mTECs) play a privileged role in this process by their unique capacity to express a diverse range of self-antigens. This gene expression program is controlled in part by the transcription factor Aire (Autoimmune Regulator). mTECs control not only the deletion of potentially hazardous T-cells but also the generation of natural regulatory T cells (nTregs), which are essential for maintaining T-cell tolerance in the periphery. Importantly, defects in mTEC development or function lead to autoimmune disorders. Although the importance of mTECs in the immune system is well recognized, the molecular and cellular mechanisms responsible for their differentiation in the thymus remain unsolved. We aim at investigating the genetic programs mechanisms that sustain mTEC differentiation as well as the molecular and cellular mechanisms that drive nTreg cell generation and function.
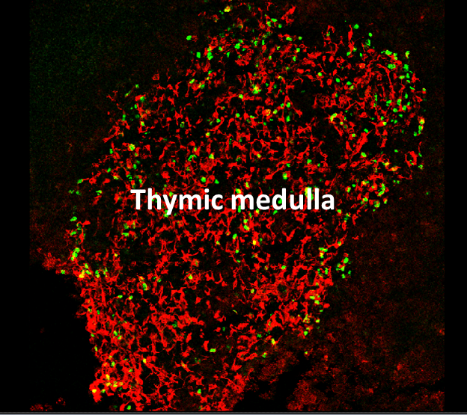
Confocal micrograph of a thymic section stained for the medulla (anti-keratin 14, red) and Aire (green)
5- Long-range interacting forces in biological systems
Biochemical interactions between bio-molecules are critical for cellular functions. The aqueous milieu in which these interactions normally take place theoretically opposes an electrostatic barrier, thus predicting a significant restrain in terms of interaction/reaction efficiency.
What are the physical forces that drive interaction between biological players with high specificity (in space, order and time)? With the groups led by Pr. M. PETTINI/Center for Theoretical Physics (CPT) and D. MARGUET/ CIML, we test the hypothesis that electro-dynamic forces partly sustain the catalytic power of cellular machines and long-range coherence in biological systems.